|
A Deep Learning Approach for Therapeutic Effectiveness Estimation of FDA Approved Anti-Viral Drug Groups by Virus Genome Sequence Analysis
By Stefano Sartori MSc May 2020 office@sartori-software.com
Scientific proofreading by Eugenio Filippi MBA MSc
Keywords Artificial Intelligence, Deep Learning, Deep Artificial Neural Network, Virus Genome, FASTA file format, Therapeutic Investigation, Anti-Viral Drug Groups, Anti-Viral Therapy, SARS-CoV-2, HBV, HCV, HIV, HPV RSV, Human Influenza Virus MERS-CoV, Ebolavirus, Acyclic nucleoside phosphonate analogues, Entry inhibitors, HCV NS5A and NS5B inhibitors, Influenza virus inhibitors, Integrase inhibitors, Interferons, immunostimulators, oligonucleotides, and antimitotic inhibitors, NNRTIs, NRTIs, Nucleoside analogues, Protease inhibitors
Abstract
This work demonstrates that a deep artificial neural network can learn to map viral genome sequences onto correlated anti-viral drug groups. Once trained, such a deep artificial neural network is able to estimate the therapeutic effectiveness of a defined drug group for novel/emerging viruses such as SARS-CoV-2, responsible of the COVID-19 pandemic humanity is currently facing.
This was achieved by training and testing different artificial neural network configurations with viral genome sequences of the following viruses: HBV Hepatitis B Virus, HCV Hepatitis C Virus, HIV Human Immunodeficiency Virus, HPV Human Papilloma Virus, Human Influenza Virus, RSV Respiratory-Syncytial-Virus.
The best performing artificial neural network was then chosen to estimate the therapeutic effectiveness of approved drug groups against viruses unknown to the network, namely Ebolavirus, MERS-CoV Middle East Respiratory Syndrome-Related Coronavirus, SARS-CoV-2 Severe Acute Respiratory Syndrome Coronavirus 2.
For actual drugs in the identified drug groups, scientific studies demonstrating their possible anti-viral capabilities for defined viruses have been published. By comparing the published results against estimated effectiveness of the different drugs for treatment of new, untested viruses, derived by using the artificial neural network, this work shows that an aid for therapeutic investigations for novel/emerging viruses like MERS-CoV and SARS-CoV-2 becomes available.
Table of Contents
2. Deep Learning and Artificial Neural Networks 3. Identifying FDA Approved Anti-Viral Drug Groups 4. Gathering the Virus Genome Sequences in Digital Format 5. Artificial Neural Network Setup, Training and Test Datasets 5.1 Neural Network Input Setup 5.2 Neural Network Output Setup 5.3 Neural Network Architecture 5.4 Training and Test Datasets 6. Parallel Training of Multiple Networks 7.3 Training and Test Error Analysis by Output
1. IntroductionThe constant improvement of computing power [1] and the ever-increasing availability of information in digital format [2], has led to a widespread application of deep learning algorithms/ artificial neural networks to a broad range of tasks, such as medical image processing [3], classification of medical images and illustrations [4] etc.
Since complete viral genome sequences are available in digital format [5][6] and given the knowledge of FDA (US Food and Drug Administration) approved anti-viral drug groups for treatment of defined viruses [7], it should be possible to train an artificial neural network which estimates the therapeutic effectiveness of approved anti-viral drug groups for novel/emerging viruses via genome sequence analysis.
In this publication, the following will be elaborated:
· Gathering the necessary information for training, test and estimation
· Set up an artificial neural network which takes virus genome sequences as input and correlated anti-viral drug group effectiveness as output.
· Train the artificial neural network with defined virus genome sequences and correlated anti-viral drug group effectiveness.
· Let the artificial neural network estimate the therapeutic effectiveness of FDA approved anti-viral drug groups for novel/emerging viruses outside the training and test data sets.
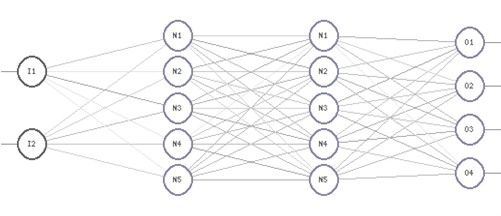
2. Deep Learning and Artificial Neural NetworksArtificial neural networks [8] can be described in general terms as systems, with the task of mapping input information to output information by generalizing potentially existing underlying rules. The architecture of an artificial neural network is defined by the task the network is required to perform. In technical terms, artificial neural networks consist of an input layer, one to many hidden layers and an output layer. Deep artificial neural networks have more than one hidden layer; the training process of such networks is called deep learning accordingly. Each network layer contains nodes, called neurons N which are connected to neurons in the previous layer through synapses. The computation flows from the network input I to its output O with each neuron value computed via the synapses as the weighted sum of the neuron values of the previous layer.
Deep learning with artificial neural networks is accomplished via supervised training [9]. During the training process, an artificial neural network is fed with actual information input-output pairs - for every input the network computes the output. The difference between actual and computed output represents the network error, which in turn is used to adjust the network parameters. Each training information input-output pair is fed through the network multiple times, the network parameters (synapse weights) are adjusted each time and over again in order to minimize the network error. The parameter adjustments correspond to the learning process of the artificial neural network, one training information feeding cycle is called “training epoch”.
The artificial neural network error [10] (the difference between computed- and actual output), is measured over the training epochs, if it decreases, the artificial neural network is said to be learning.
To find the best performing artificial neural network for a given task, multiple artificial neural networks with different setup parameters are trained with the same data, the best performing artificial neural network is then chosen for the designed task. The network with the lowest test and training error is said to be the best performing network.
Deep learning and artificial neural networks keywords for further reading: Supervised Learning, Feature Scaling, Activation Functions, Backpropagation, Stochastic Gradient Descent (SGD), Weight Initialisation, Weight Decay, Sparsity/Density, Cross Validation.
3. Identifying FDA Approved Anti-Viral Drug GroupsThe publication available under https://cmr.asm.org/content/29/3/695 [7] gives an overview of the FDA approved antiviral drugs over the past 50 years, which can be summarized by virus and drug group as follows.
HBV Hepatitis B Virus HCV Hepatitis C Virus HIV Human Immunodeficiency Virus HPV Human Papilloma Virus Human Influenza Virus RSV Respiratory-Syncytial-Virus
4. Gathering the Virus Genome Sequences in Digital FormatComplete viral genome sequences are available in the FASTA file format from the United States National Center for Biotechnology Information (NCBI) under the following link: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ [5] Viral genome sequences are represented in the FASTA file format [6] as a sequence of the characters A,C,G,T which represents the nucleic acid bases: adenine, cytosine, guanine and thymine respectively.
Viral genome sequences length in nucleic acid base count [5]:
Eligible viruses for training and test have been chosen by their genome sequence size, since the genome length has a direct impact on the network size, which in turn heavily influences training time on the chosen hardware.
5. Artificial Neural Network Setup, Training and Test Datasets5.1 Neural Network Input SetupThe viral genome sequence [5] has to be transformed to numerical values which the artificial neural network is able to process. For each nucleic acid base, represented by one of the four characters A,C,G,T in the viral genome sequence [6], a number of 4 neural network inputs must be setup [9], thus, the total number of inputs depends on the maximum viral genome sequence length the network is capable of processing. Each nucleic base in the viral genome sequence is mapped to a group of four inputs as follows:
Since the task is to estimate the therapeutic effectiveness of approved drug groups for Ebolavirus, MERS-CoV and SARS-CoV-2, the network is designed to accept a maximum viral genome sequence length of 32,000 nucleic acid bases. This results in 4 x 32,000 = 128,000 inputs for the artificial neural network.
5.2 Neural Network Output SetupThe neural network output setup is given by the drug groups [7] which the network should map onto the input viral genome sequence [5].
5.3 Neural Network ArchitectureGiven the conditions outlined in neural network input and output setup sections, the following artificial neural network architecture has been chosen:
Input layer with 128,000 neurons for the viral genome sequence, 3 hidden layers with 20,000 neurons for each layer, output layer with 10 neurons representing the drug groups.
Activation functions have a random uniform distribution for each neuron in the network: Softplus, Logistic, Hyperbolic Tangent and ReLU. [8][9]
Input and output data feature rescaling [8][9] in interval [-1,1]
Since a fully connected network [8] with this architecture imply prohibitive computing costs, the following synapse density factors have been setup:
Layer 1 (Input) to Layer 2: 0.00008 Layer 2 to Layer3: 0.0001 Layer 3 to Layer 4: 0.0001 Layer 4 to Layer 5 (Output): 0.1
The density factors have been chosen in order to connect each neuron in a layer with at least 2 neurons from the previous layer.
Learning related hyper parameters [8]:
Weight Initialisation Method: Xavier Normal Weight Decay: 0.1 Initial Learning Rate: 0.0001 Initial Momentum: 0.00001 Batch Size: 1 (full online)
The learning related hyper parameters have
been chosen to avoid network overfitting on the training data set. 5.4 Training and Test DatasetsAs training and test dataset, 100 complete genome sequences [5] for each virus species HBV, HCV, HIV, HPV, Human Influenza and RSV have been mapped onto the FDA approved drug group [7], resulting in a total number of 600 complete genome sequences. 80% and 20% [9] of the genome sequences were set up for training and test dataset respectively. The dataset used to train the artificial neural network can be generalized as follows:
Input I1 - I128,000 Virus genome sequence input from FASTA files, nucleic acid bases loaded into the artificial neural network input.
Output O1 - O10 Drug Groups/Effectiveness 6. Parallel Training of Multiple Networks
6.1 MethodologyIn order to find the best performing artificial neural network, 4 networks with uniform random variations in their hyper parameters have been trained in each training batch in parallel. Uniform random variations of hyper parameters have been chosen over the permutation of hyper parameters in order to control the maximum number of networks to be trained in parallel. To optimize the performance, the number of networks to be trained in parallel has been restricted to match the number of CPUs/CPU cores of the computer used. The RAM utilisation per network was about 2.7 GB while holding the network setup, the complete training and the test dataset in memory.
The training process for each batch, containing 4 artificial neural networks, took between 6 and 12 hours computing time. The fluctuation of the training time depended on the chosen network hyper parameters and learning behaviour over the training epochs.
20 training batches with a total number of 80 different neural networks have been executed over the period of a week resulting in the identification of the best performing network, namely the one with the lowest training and test error.
6.2 HardwareIntel® Xeon® E3-1246v3 4 Cores 3.5GHz Hyper-Threading 16 GB RAM 1 TB HDD (RAID 1) + 250 GB SSD (RAID 1)
6.3 SoftwareSSWAI by sartori-software.com: neural network management, setup, training validation and production application, written in PHP 7.x
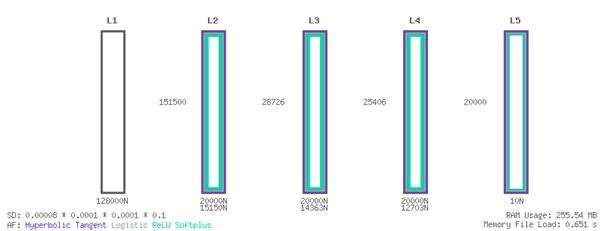
7. Best Performing NetworkThe parallel training process described in the previous section, yielded the following best performing artificial neural network, identified as the one with the lowest training and test error, after 148 training epochs.
7.1 Architecture
Layer L1 Input; Virus Genome Sequence; 128,000 Neurons Layer L2 15,150 Neurons; 151,500 Synapses to L1 Layer L3 14,363 Neurons; 28,726 Synapses to L2 Layer L4 12,703 Neurons; 25,406 Synapses to L3 Layer L5 Output; Drug Group; 10 Neurons; 20,000 Synapses to L4
7.2 Error Function
X: Epochs N=148 Y: Network Error: Sum of squared error over all outputs divided by training and test record count respectively
The training has been actively stopped at Epoch 148, since no significant decrease of the network error could be observed. In this specific case, about half the number of epochs used, would have been sufficient to reduce both the training and test error to an acceptable level.
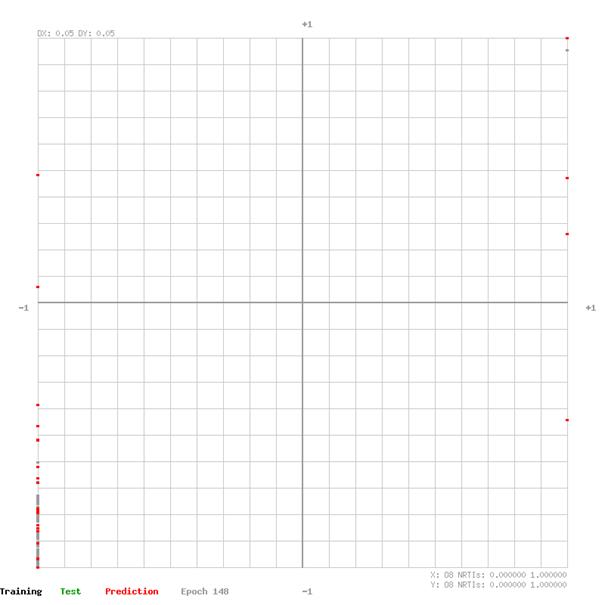
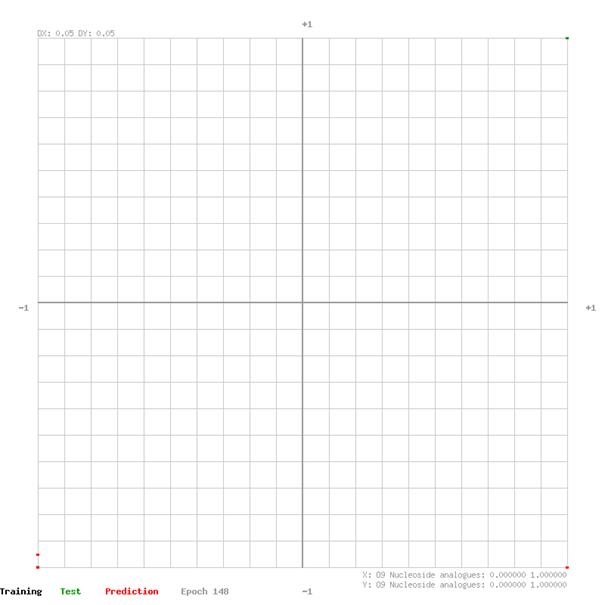
7.3 Training and Test Error Analysis by Output
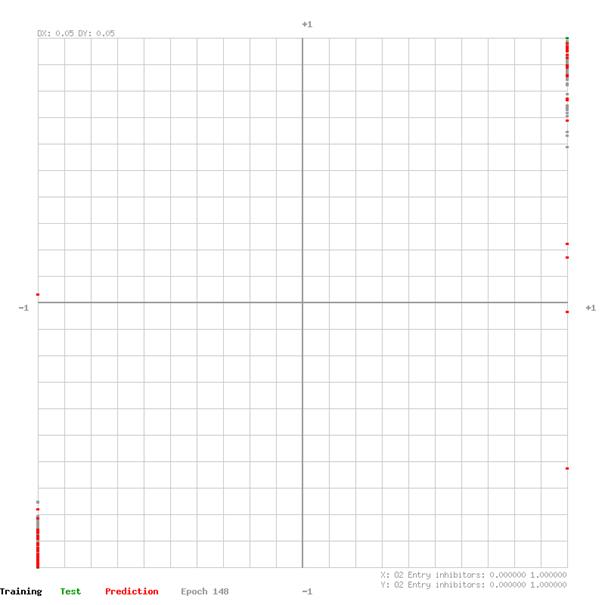
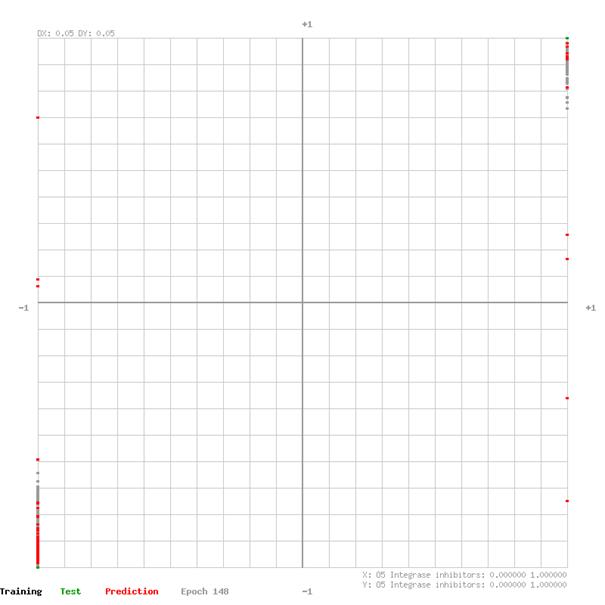
The network output of training and test data sets are plotted in the graphs on the following pages. The graph XY domain [-1,1] is given from the applied feature rescaling before feeding the data to the network. The closer the data points are to the top-right- and bottom-left- corner of the graph the better the network estimates the effectiveness of the drug group for a defined virus.
O1 Acyclic nucleoside phosphonate analogues
O2 Entry inhibitors
O3 HCV NS5A and NS5B inhibitors
O4 Influenza virus inhibitors
O5 Integrase inhibitors
O6 Interferons, immunostimulators, oligonucleotides, and antimitotic inhibitors
O7 NNRTIs
O8 NRTIs
O9 Nucleoside analogues
O10 Protease inhibitors
7. Ebolavirus Estimation443 complete Ebolavirus virus genome sequences have been gathered from the United States National Center for Biotechnology Information under the following link: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ [5]
The 443 complete virus genome sequences have then been fed into the best performing artificial neural network identified. Since the network was trained to map a drug group effectiveness for a defined virus [7], onto its viral genome [5], with 0 deemed not effective and 1 deemed effective, the output values give therapeutic effectiveness estimation for the defined drug group. The table below shows the median of the output values by drug group, computed by the network for the 443 complete Ebolavirus virus genome sequences [5], where greater values up to 1 represent greater effectiveness estimate, and smaller values close to 0 represent low or no effectiveness estimate.
8. MERS-CoV Estimation257 complete MERS-CoV virus genome sequences have been gathered from the United States National Center for Biotechnology Information under the following link: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ [5] The 257 complete virus genome sequences have then been fed into the best performing artificial neural network identified. Since the network was trained to map a drug group effectiveness for a defined virus [7], onto its viral genome [5], with 0 deemed not effective and 1 deemed effective, the output values give therapeutic effectiveness estimation for the defined drug group. The table below shows the median of the output values by drug group, computed by the network for the 257 complete MERS-CoV virus genome sequences [5], where greater values up to 1 represent greater effectiveness estimate, and smaller values close to 0 represent low or no effectiveness estimate.
9. SARS-CoV-2 Estimation941 complete SARS-CoV-2 genome sequences have been gathered from the United States National Center for Biotechnology Information under the following link: https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/ [5] The 941 complete virus genome sequences have then been fed into the best performing artificial neural network identified. Since the network was trained to map a drug group effectiveness for a defined virus [7], onto its viral genome [5], with 0 deemed not effective and 1 deemed effective, the output values give therapeutic effectiveness estimation for the defined drug group. The table below shows the median of the output values by drug group, computed by the network for the 941 complete SARS-CoV-2 genome sequences [5], where greater values up to 1 represent greater effectiveness estimate, and smaller values close to 0 represent low or no effectiveness estimate.
10. ConclusionsThe deep learning approach to estimate therapeutic effectiveness of anti-viral drug groups described in this publication provided the following results to focus on:
Matching the identified drug groups by using the publication available under https://cmr.asm.org/content/29/3/695 to a selection of actual drugs and available publications yields the following table:
The neural network was excluded from having access to any of the Ebolavirus, MERS-CoV and SARS-CoV-2 genome sequences during the training and test process.
The estimated results therefore indicate clearly that it is possible to train artificial neural networks on known viral genomes and correlated therapies to aid therapeutic investigations for novel/emerging viruses like MERS-CoV and SARS-CoV-2.
With time, computing power will still increase [1] and more complete genome sequences are expected to be available in digital format [2], making it possible to train more accurately artificial neural networks [8] for genomic sequence analysis tasks.
As next step it is planned to apply the methodology described to viral vaccines. Considering the mechanism of action of vaccines [11], it seems unlikely that an artificial neural network with reasonable low error can be trained. On the other hand, if an acceptable performing artificial neural network is found, it could be an indication of possible viral vaccine cross-protection.
The methodology described could also be used
to investigate anti-bacterial therapies, though training an artificial neural
network on bacteria genomes will require significantly more computing power or
training time, as bacterial genomes are orders of magnitudes larger than viral
genomes [12]. 11. References[1] Moores’Law - Wikipedia Article https://en.wikipedia.org/wiki/Moore%27s_law
[2] Data generated by humanity - Forbes Article
[3] Deep Learning for Medical Image Processing - Science Direct Article https://www.sciencedirect.com/science/article/pii/S0939388918301181
[4] Artificial Neural Networks for Medical Image Classification - NCBI Publication https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4699299/
[5] Online Virus Genome Sequences https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#/
[6] Definition of the FASTA format - Wikipedia Article https://en.wikipedia.org/wiki/FASTA_format
[7] FDA Approved Anti-Viral Drug Groups - Clinical Microbiology Reviews Publication https://cmr.asm.org/content/29/3/695
[8] Introduction to Artificial Neural Networks - Article https://towardsdatascience.com/introduction-to-artificial-neural-networks-ann-1aea15775ef9
[9] Supervised Training -Medium Article
[10] Network Error, Error Function - NCBI Publication https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5009026/
[11] How Vaccines Work - World Health Organization Publication
[12] Genome Size - Science Direct Page https://www.sciencedirect.com/topics/medicine-and-dentistry/genome-size 12. About the AuthorStefano Sartori, born 1973 in Italy
1992 Technical high school degree in electronics and matriculation University of Bologna, Italy, faculty of physics.
1995-1996 - 12 months exchange Student at the Technical University Vienna, Austria - ERASMUS-SOCRATES Project. Specialization in Biophysics, radiation protection and dosimetry.
1998 Degree in Physics (MSc) Thesis: Implementation of a computerized management system for the controlled disposal of radioactive waste deriving from the medical use of radionuclides of storage facility identified by: IAEA "TECDOC-775, HANDLING, TREATMENT, CONDITIONING AND STORAGE OF BIOLOGICAL RADIOACTIVE WASTES International Atomic Energy Agency (IAEA)"
1998-2020 Owner of sartori-software.com - Software Company with core business in application development for the pharmaceutical industry. Continuous improvement of IT skills and software development technologies, specialization in Deep Learning and Artificial Neural Networks.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||